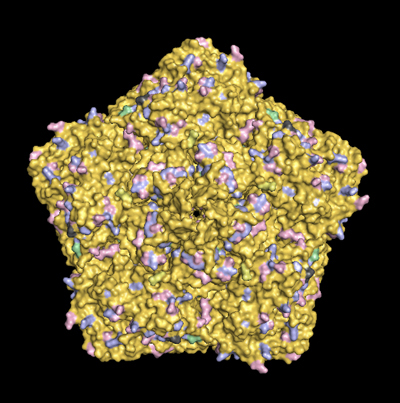
This page compares the capsid surface residues of CVB3 strains currently being studied. It is intended to facilitate distance discussions with my collaborators and details are currently sparse. Results of our studies with strains CVB3/28, CVB3/RD and CVB3/RDVa can be read at PMID: 21270163.
In the second image ( bottom of the page), mouse over CAR or DAF to see where one of these proteins can bind to one of the 5 available sites on the pentamer.
In the table below, click on the virus strain to display a base image (left column) then mouse over the strain capsid you want to overlay (right column).
The color scheme shows glu/asp (pink), lys/arg (pale blue), his (pale green). Key differences among the strains are emphasized by darker colors. Dark gray is used for non-charged subsitutions.
Stability and capsid diffrences for CVB3/28BS, /28CC, and /28N have been reported (J. Gen. Virol. 97:60-68). CVB3/28N (half-life at 37C is 17hr) differs from CVB3/28 (half-life at 37C is 7hr) in only three of the capsid protein amino acids. The published results show that CVB3/28 is one of very few enteroviruses with leucine at residue 92 of Vp1, and that mutation of the leucine to valine is able to stabilize the virus (half-life is 17hr).
Images rendered in PyMol are based on 1COV.PDB [Muckelbauer, J. K., M. Kremer, I. Minor, L. Tong, A. Zlotnick, J. E. Johnson, and M. G. Rossmann. 1995. Structure determination of coxsackievirus B3 to 3.5 A resolution. Acta Crystallogr. D. Biol. Crystallogr. 51:871-877] and structural data for CAR binding [He, Y., P. R. Chipman, J. Howitt, C. M. Bator, M. A. Whitt, T. S. Baker, R. J. Kuhn, C. W. Anderson, P. Freimuth, and M. G. Rossmann. 2001. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nat. Struct. Biol. 8:874-878] and DAF binding [Yoder JD, Cifuente JO, Pan J, Bergelson JM, Hafenstein S. J Virol. 2012 Sep 12. [Epub ahead of print]] |